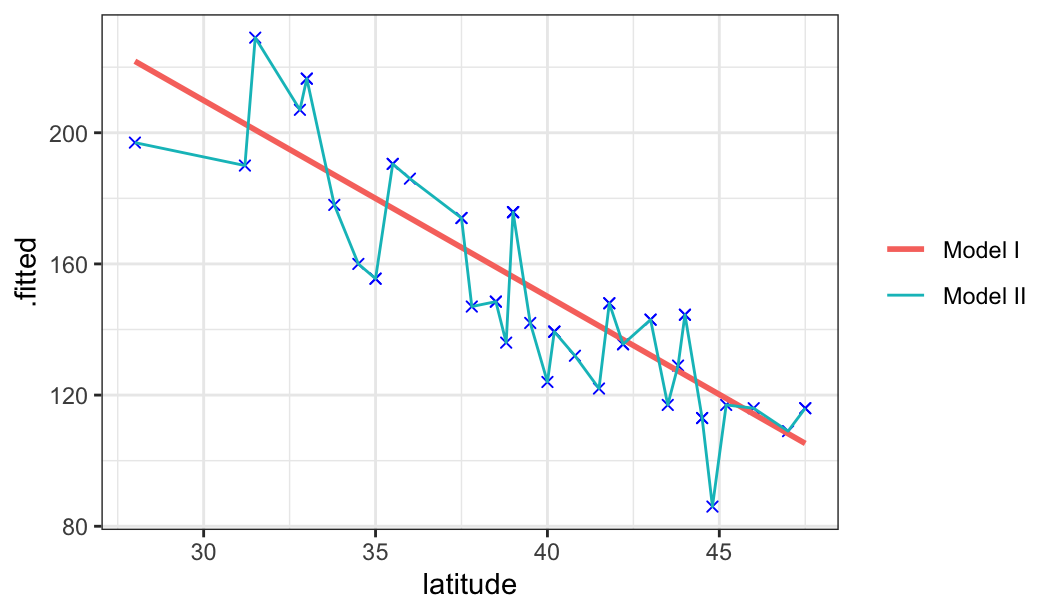
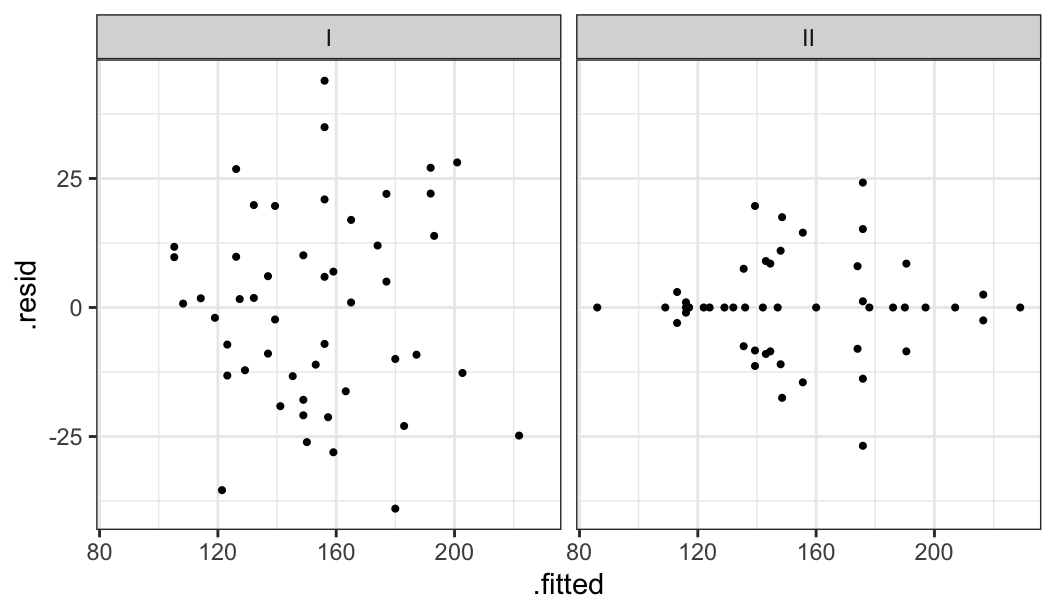
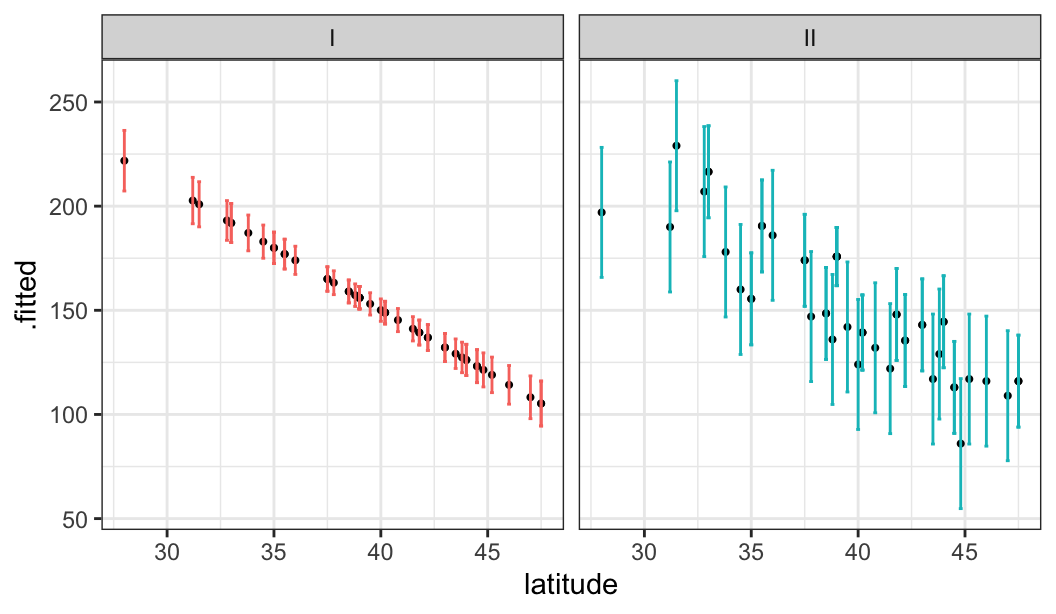
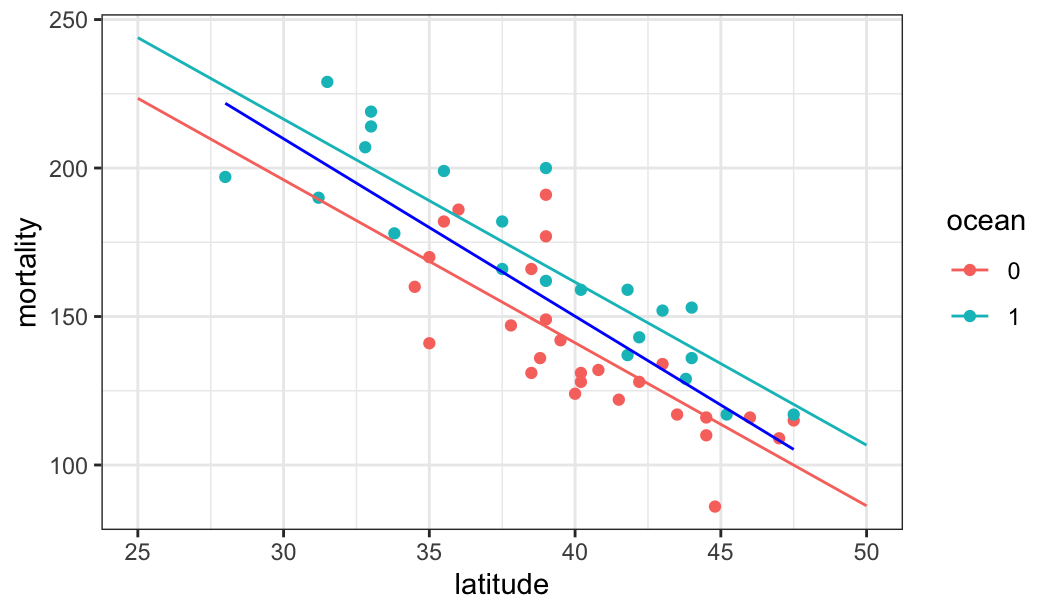
glimpse(mordat)Rows: 49
Columns: 6
$ state <chr> "Alabama", "Arizona", "Arkansas", "California", "Colorado", …
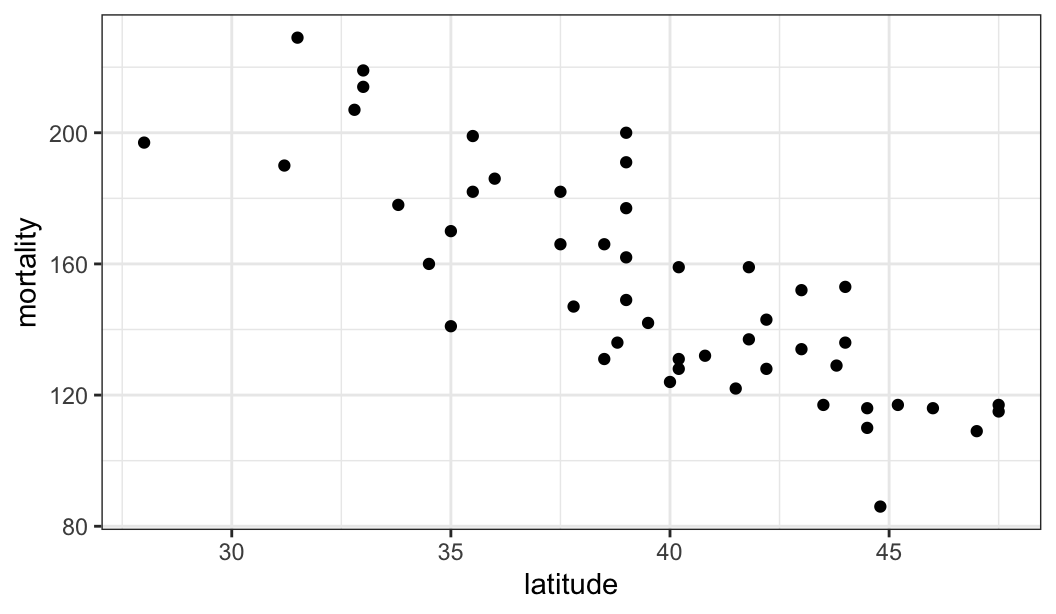
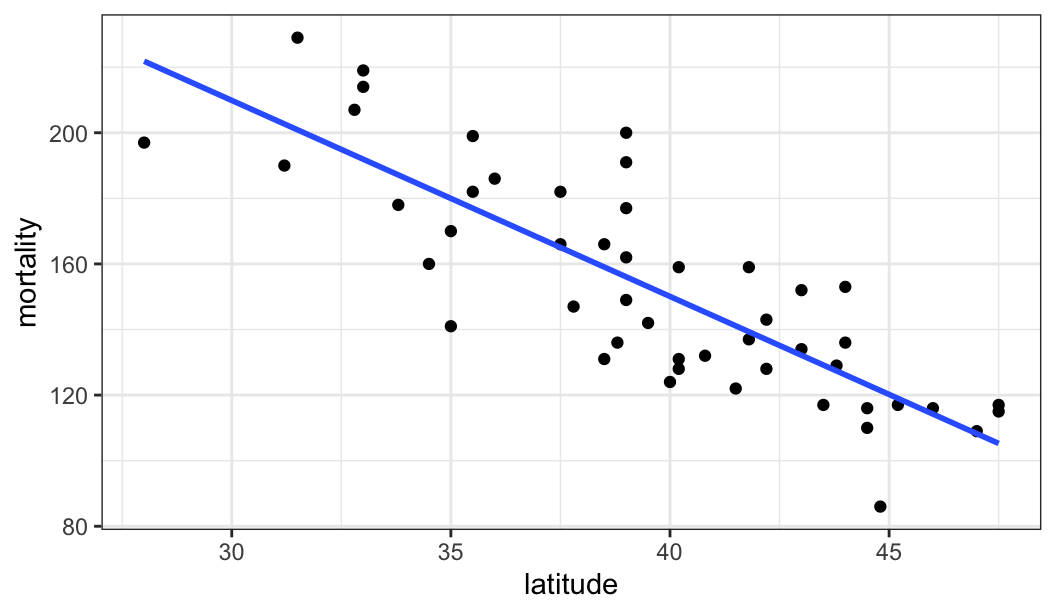
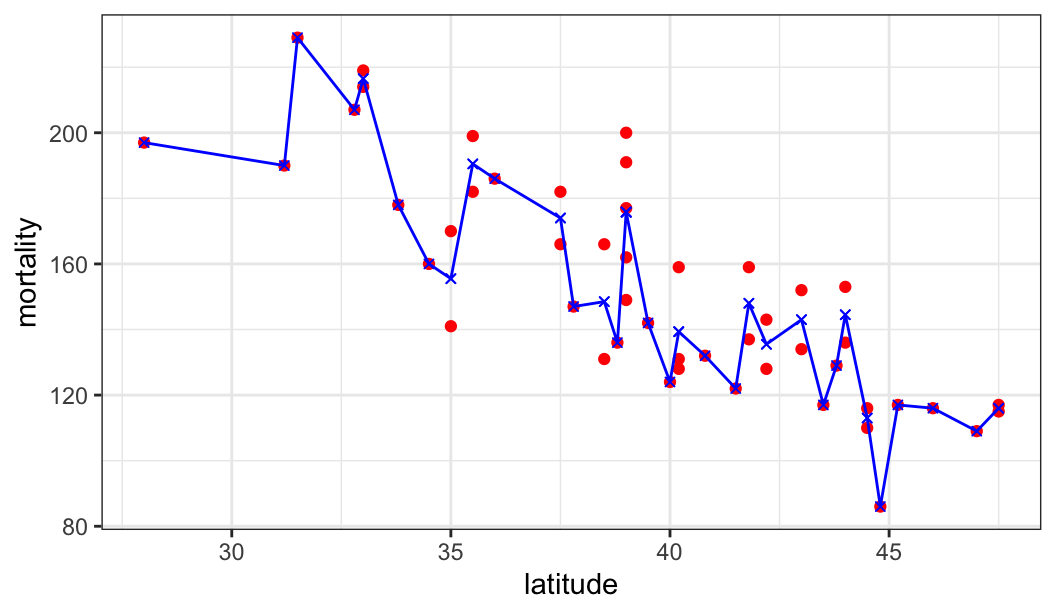
$ mortality <dbl> 219, 160, 170, 182, 149, 159, 200, 177, 197, 214, 116, 124, …
$ latitude <dbl> 33.0, 34.5, 35.0, 37.5, 39.0, 41.8, 39.0, 39.0, 28.0, 33.0, …
$ longitude <dbl> 87.0, 112.0, 92.5, 119.5, 105.5, 72.8, 75.5, 77.0, 82.0, 83.…
$ pop <dbl> 3.46, 1.61, 1.96, 18.60, 1.97, 2.83, 0.50, 0.76, 5.80, 4.36,…
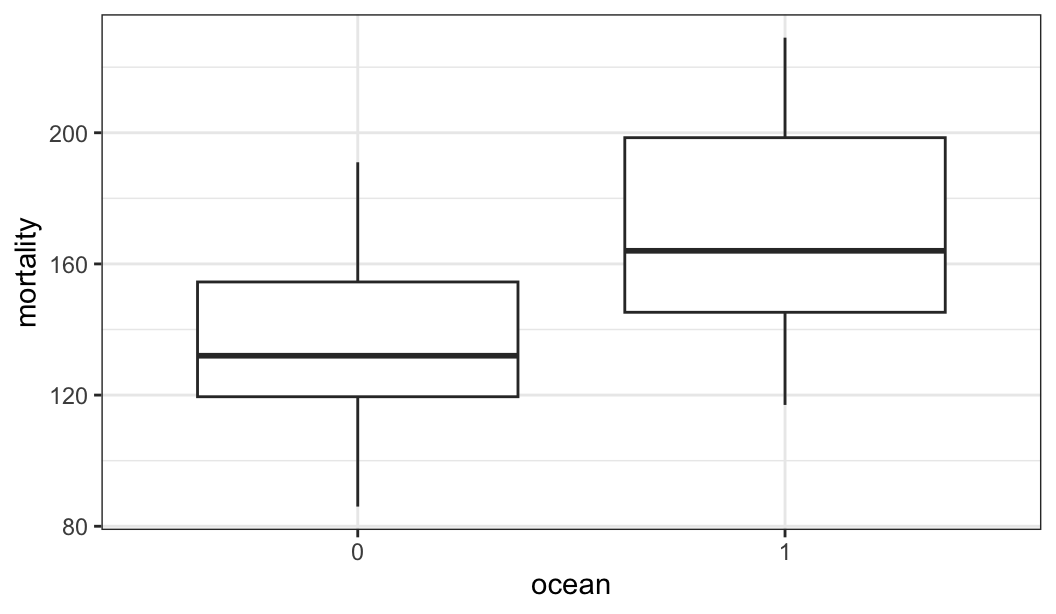
$ ocean <fct> 1, 0, 0, 1, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, …