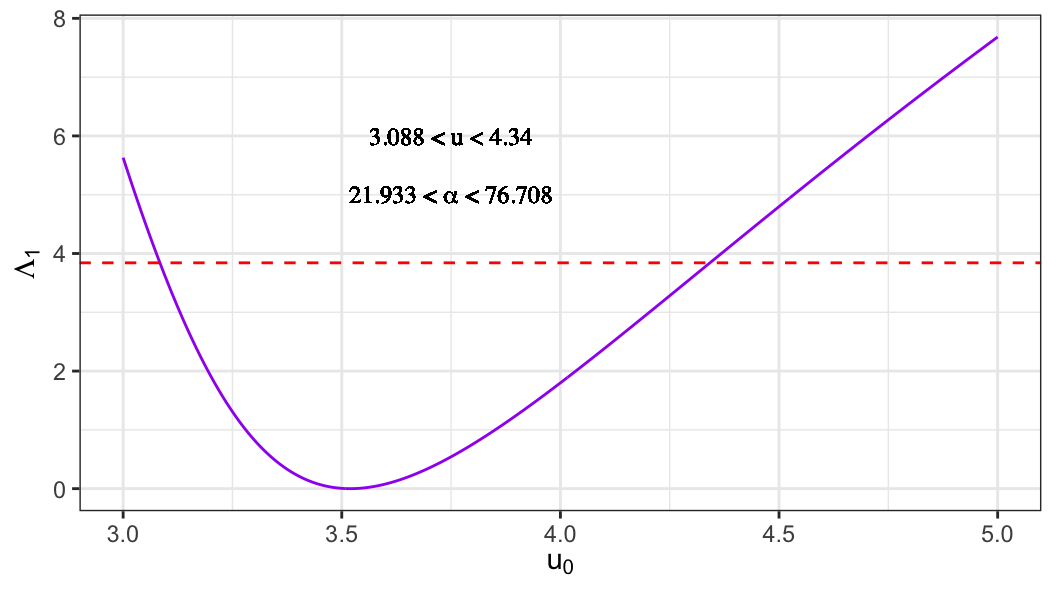
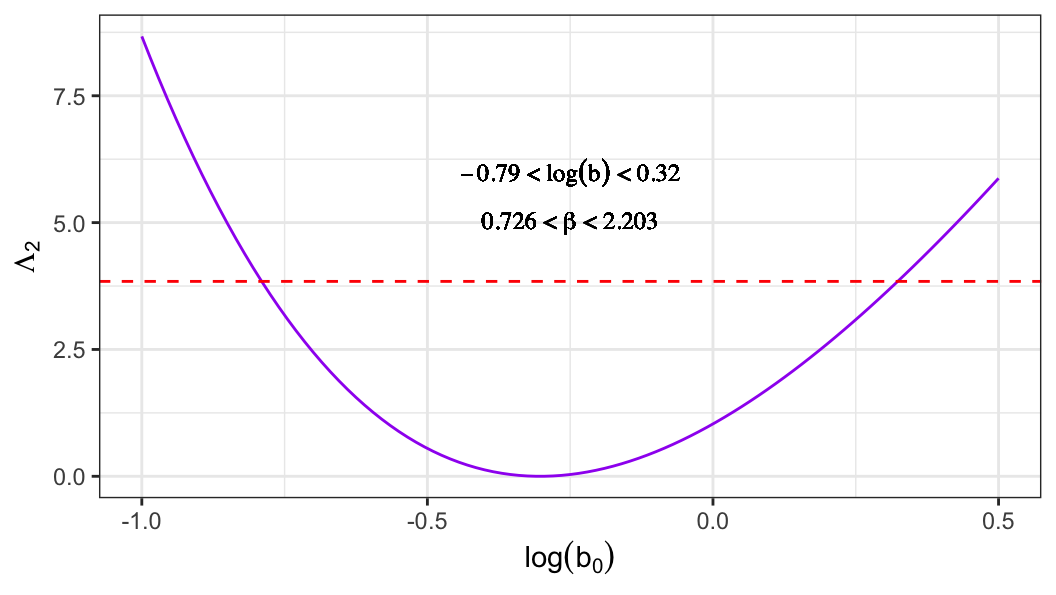
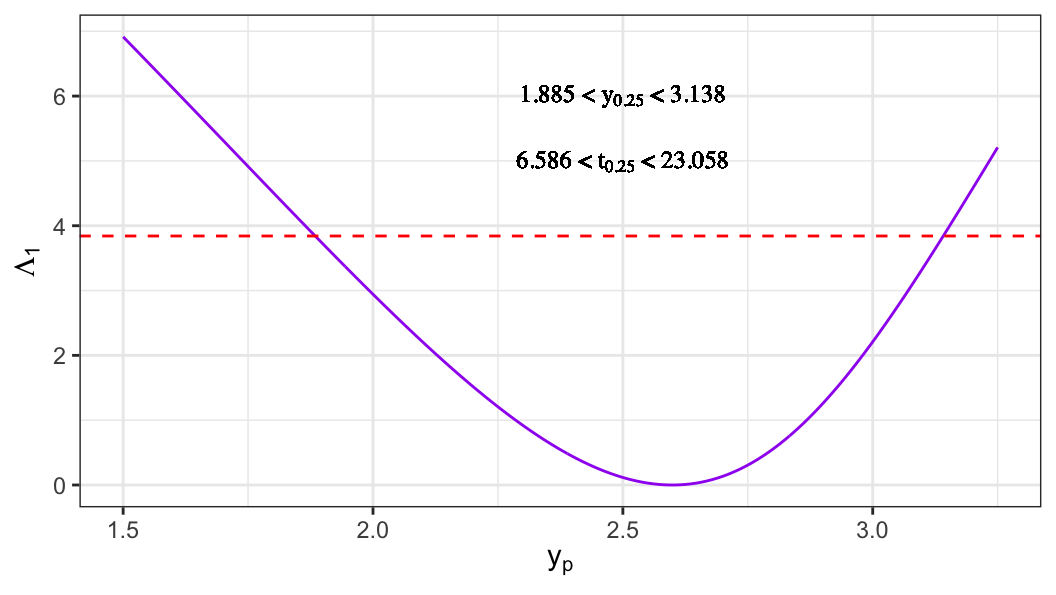
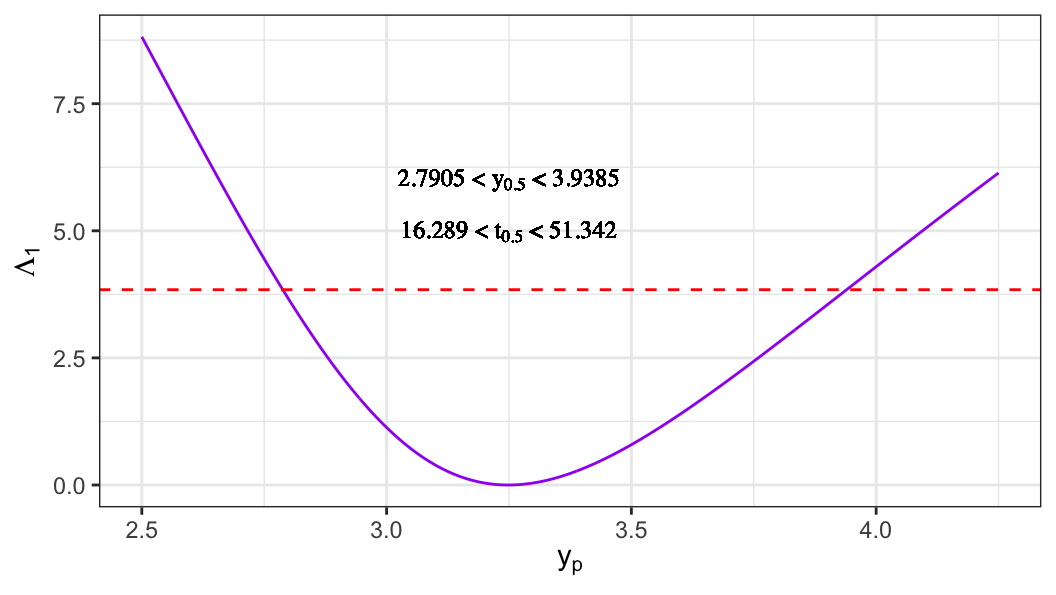
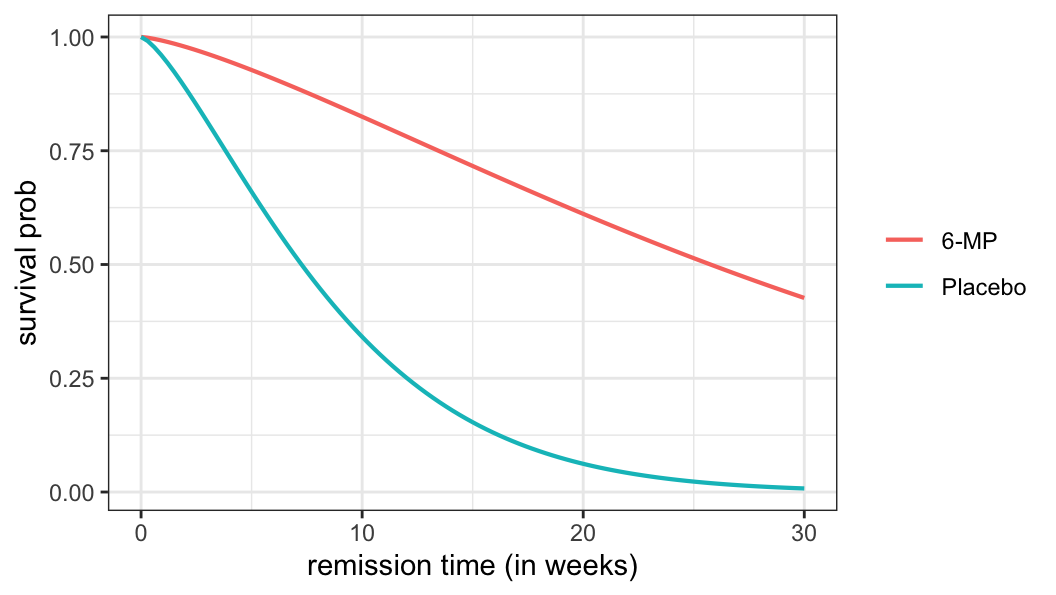
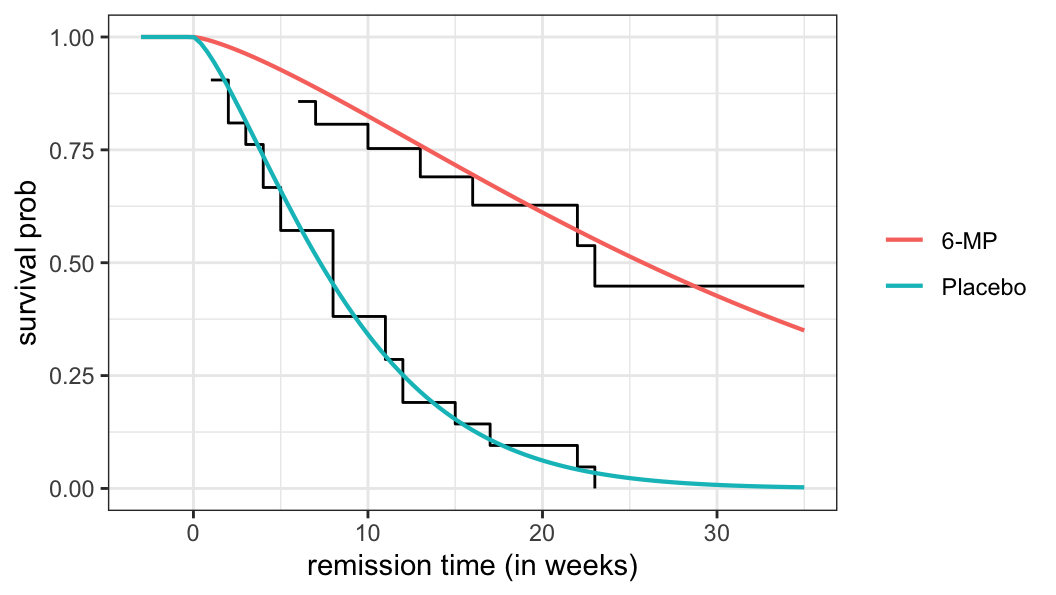
- For extreme-value distribution
\[\begin{align*}
\frac{\partial^2 \ell(u, b)}{\partial u^2}
& = \frac{1}{b^2} \sum_{i=1}^n\bigg[\delta_i\,\frac{\partial^2 \log f_0(z_i)}{\partial z_i^2} + (1-\delta_i)\, \frac{\partial^2 \log S_0(z_i)}{\partial z_i^2}\bigg] =-\frac{1}{b^2} \sum_{i=1}^n e^{z_i}
\end{align*}\] where \[\begin{align*}
\frac{\partial^2 \log f_0(z)}{\partial z^2} & = \frac{\partial}{\partial z} \big\{1 - e^z\big\} = -e^z=
\frac{\partial^2 \log S_0(z)}{\partial z^2}
\end{align*}\]
\[\begin{align}
{\color{purple} \frac{\partial^2 \ell(u, b)}{\partial b^2} \bigg\vert_{u=\hat u, \,b=\hat b}=-\frac{1}{\hat b^2} \sum_{i=1}^n e^{\hat z_i}=-\frac{r}{\hat b^2}}
\end{align}\]
From Section 5.1.4, it can be shown that the Hessian matrix will be
\[\begin{align*}
\frac{\partial^2 \ell(u, b)}{\partial b^2} & = \frac{r}{b^2} +\frac{2}{b^2} \sum_{i=1}^nz_i\bigg[\delta_i\,\frac{\partial \log f_0(z_i)}{\partial z_i} + (1-\delta_i)\, \frac{\partial \log S_0(z_i)}{\partial z_i}\bigg]\\[.25em]
& \;\;\;+\frac{1}{b^2}\sum_{i=1}^nz_i^2\bigg[\delta_i\,\frac{\partial^2 \log f_0(z_i)}{\partial z_i^2} + (1-\delta_i)\, \frac{\partial^2 \log S_0(z_i)}{\partial z_i^2}\bigg]\\[.25em]
& = \frac{r}{b^2} +\frac{2}{b^2} \sum_{i=1}^nz_i \big[\delta_i(1-e^{z_i}) -(1-\delta_i)e^{z_i}\big] -\frac{1}{b^2}\sum_{i=1}^nz_i^2 e^{z_i} \\[.25em]
&=\frac{r}{b^2} +\frac{2}{b^2} \sum_{i=1}^nz_i\big[\delta_i-e^{z_i}\big] -\frac{1}{b^2}\sum_{i=1}^nz_i^2 e^{z_i}
\end{align*}\]
\[\begin{align}
{\color{purple} \frac{\partial^2 \ell(u, b)}{\partial b^2} \bigg\vert_{u=\hat u,\,b=\hat b}= -\frac{r}{\hat b^2}-\frac{1}{\hat b^2}\sum_{i=1}^n\hat z_i^2 e^{\hat z_i}}
\end{align}\]
\[\begin{align*}
\frac{\partial^2 \ell(u, b)}{\partial u\,\partial b}
& = \frac{1}{b^2} \sum_{i=1}^n\bigg[\delta_i\,\frac{\partial \log f_0(z_i)}{\partial z_i} + (1-\delta_i)\, \frac{\partial \log S_0(z_i)}{\partial z_i}\bigg]\\[.25em]
&\;\; \;\;+ \frac{1}{b^2} \sum_{i=1}^nz_i\bigg[\delta_i\,\frac{\partial^2 \log f_0(z_i)}{\partial z_i^2} + (1-\delta_i)\, \frac{\partial^2 \log S_0(z_i)}{\partial z_i^2}\bigg] \\[.25em]
& = \frac{1}{b^2} \sum_{i=1}^n\Big[\delta_i(1-e^{z_i}) - (1-\delta_i)e^{z_i}-z_ie^{z_i}\Big]\\[.25em]
& = \frac{1}{b^2} \sum_{i=1}^n\Big[\delta_i - e^{z_i}-z_ie^{z_i}\Big] = \frac{1}{b^2} \Big[r -\sum_{i=1}^n e^{z_i}-\sum_{i=1}^n z_ie^{z_i} \Big]
\end{align*}\]
\[\begin{align}
{\color{purple} \frac{\partial^2 \ell(u, b)}{\partial u\,\partial b} \bigg\vert_{u=\hat u,\,b=\hat b}= -\frac{1}{\hat b^2}\sum_{i=1}^n\hat z_i e^{\hat z_i}}
\end{align}\]